Lab Research and Projects
The Liang Laboratory focuses on integrating the power and advantages of different imaging system modalities, including those using electrons, x-rays, and visible lights, and scrutinizing how macromolecular machinery functions in various biological processes in real-time at unprecedented resolution. Capitalizing on breakthroughs in detector technology and image processing algorithms, the Liang laboratory is particularly interested in using cryo-electron microscopy (cryo-EM), the technology that won the 2017 Nobel Prize in Chemistry, as a primary tool to visualize macromolecular complexity. Cryo-EM is exceptionally well suited for molecular systems traditionally challenging for structural characterization, including membrane proteins and large and heterogeneous assemblies. Also, we employ X-ray crystallography and fluorescence light microscopy to provide complementary structural and dynamic information for challenging systems of significant biological importance.
Primary Projects
One such challenging system is the RNA synthesis machinery of a class of pathogenic and sometimes deadly non-segmented negative-sense (NNS) RNA viruses, including measles (MV), rabies (RABV), Ebola (EBOV), Marburg (MARV), and respiratory syncytial virus (RSV). RNA synthesis is central to the life cycle of NNS RNA viruses, carried out by a multi-functional RNA polymerase. The structural basis of the RNA synthesis machinery remains mostly unclear.
The Liang laboratory is devoted to understanding the structure and mechanism of the RNA synthesis machinery of RSV, the leading cause of severe respiratory tract diseases in young children and older adults, and immunocompromised in the United States and worldwide. RNA synthesis by the RdRP of RSV is essential for viral pathogenesis. The Liang laboratory aims to establish an RNA synthesis platform for RSV, elucidate how this RNA synthesis machine functions, and identify potential antiviral therapeutic targets for more effective treatment. Our immediate research goal would be to decipher the molecular architecture of the RNA synthesis machinery of these viruses using cryo-EM and x-ray crystallography. Our research could lead to the rational design of effective antiviral drugs to block RSV activity. Such drugs would substantially reduce serious RSV infections in humans and greatly improve human health.
Collaborative Projects
The Liang Laboratory seeks to exploit our structural biology (cryo-EM & x-ray crystallography) and biochemistry expertise to elucidate the molecular machinery and associated mechanisms of traditionally challenging and exciting biological systems through collaboration. We are particularly interested in interdisciplinary research in molecular cell biology, microbiology, neurobiology, and cancer biology. Please email Dr. Liang (bo.liang@emory.edu) directly for any potential collaboration suggestions. We look forward to working with you. Thank you!
Ongoing Projects
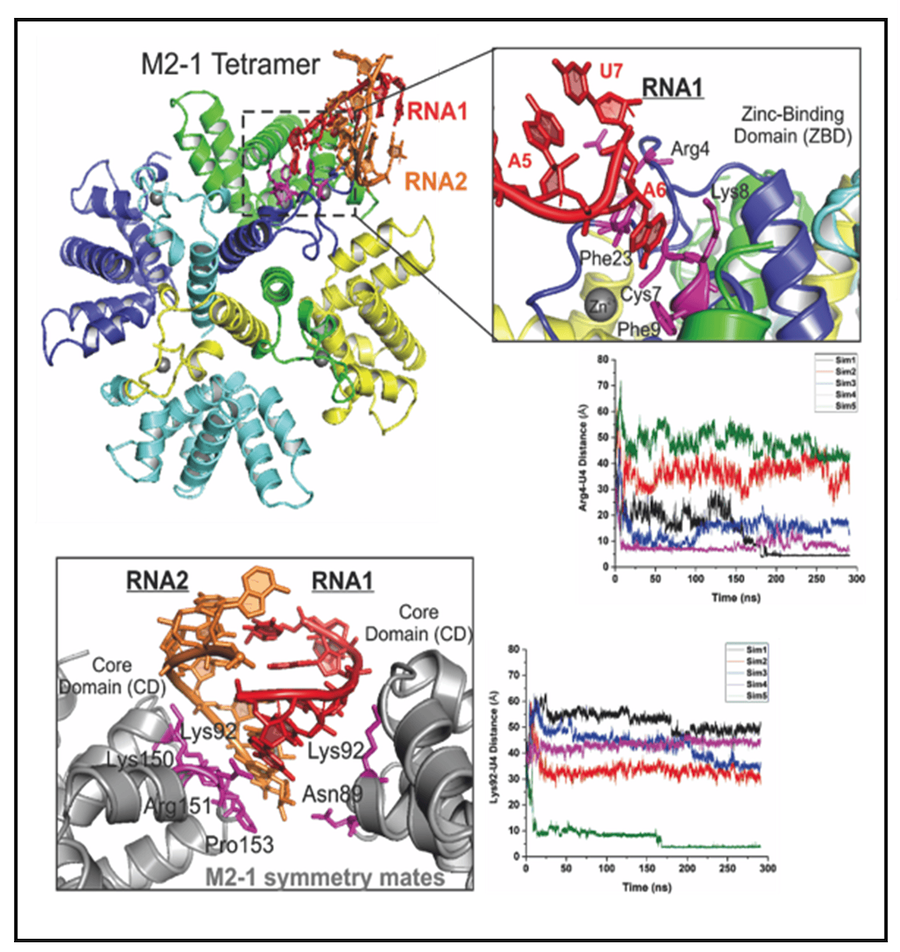
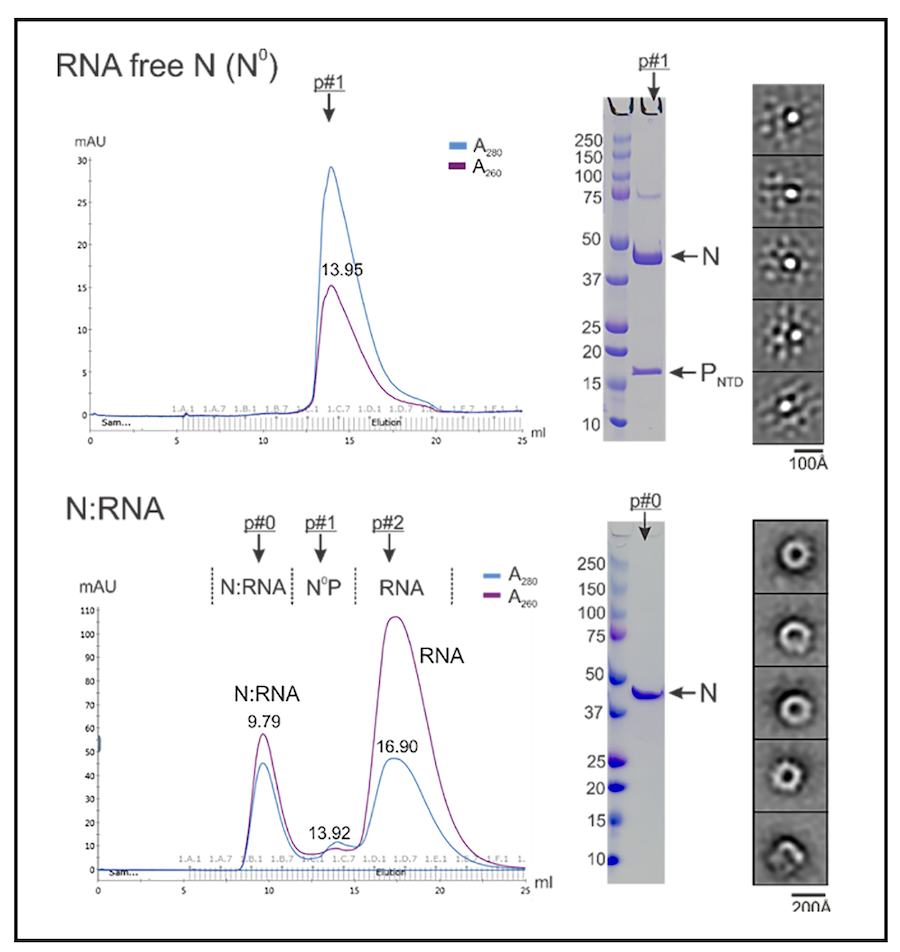
NNS RNA viruses share a common strategy of viral gene expression. NNS RNA synthesis is believed to follow the “start-stop model” of sequential and polar transcription (Fig. 1). We use RSV as a model system to delineate NNS RNA synthesis. The template for RNA synthesis is not RNA alone but rather a complex of the viral genomic RNA completely encapsidated by the viral nucleoprotein (N). This N:RNA template is copied by the viral RNA-dependent RNA polymerase (RdRP), which comprises 250 kDa large protein (L) and 27 kDa cofactor phosphoprotein (P). An additional 22kDa viral protein M2-1 is essential for full processivity in RSV. The L protein has all the enzymatic activities necessary for the transcription of the viral mRNAs, including RNA polymerization, 5’ cap addition, cap methylation, 3’ polyadenylation, and viral genome replication. Thus, L is the catalytic core of a multi-component and multi-functional RNA synthesis machine. We have successfully established protocols to prepare all key components of the RSV RNA synthesis machine. Our work on the structural and biochemical mechanisms of the RNA synthesis machinery has yielded a solid foundation for in-depth mechanistic studies.
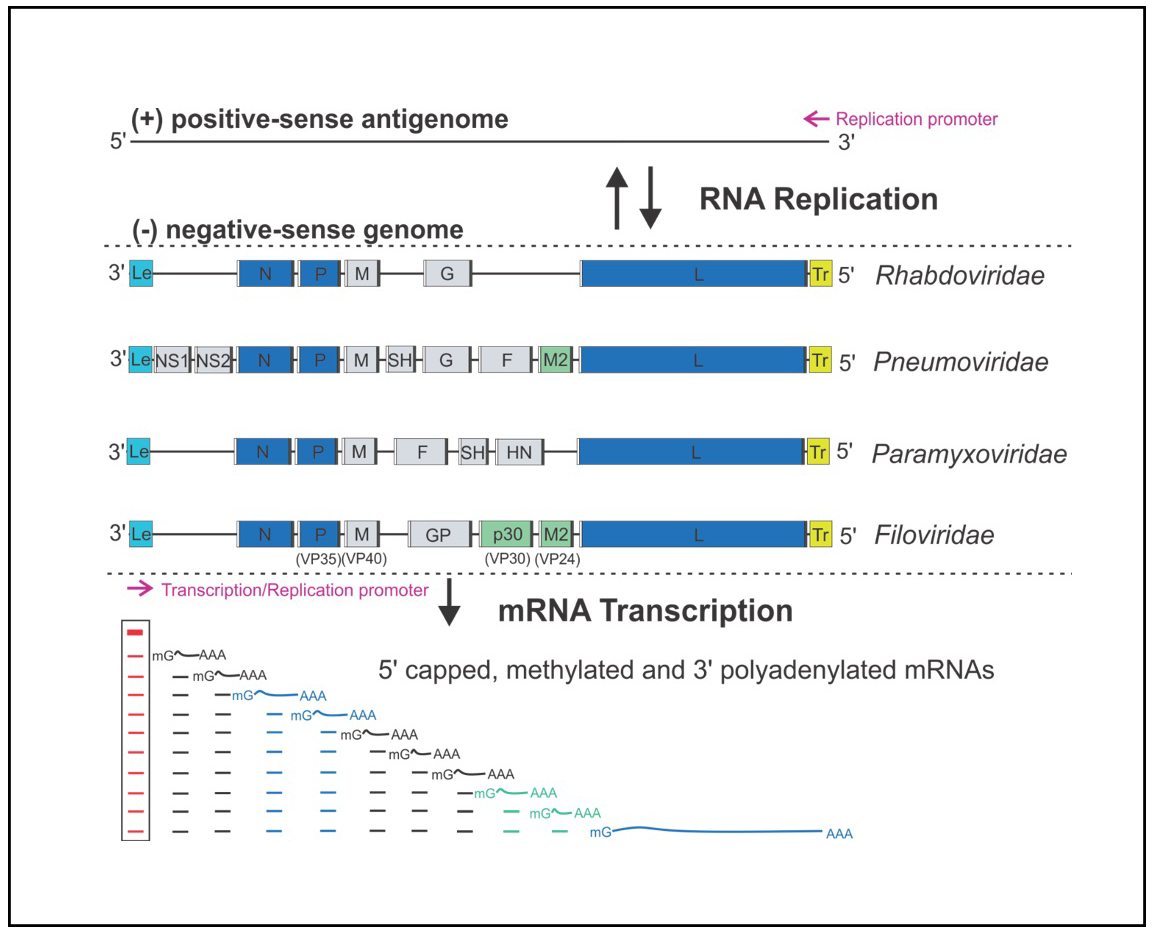 Figure 1: The genome organization and RNA synthesis of Mononegavirales. The negative-sense NNS genome is depicted from the 3′ end to the 5′ end, showing the 3′ leader (Le, cyan box), genes (gray, blue or green box) flanking with gene-start (GS, white box), and gene-end (GE, black box), and 5′ trailer (Tr, yellow box). The essential genes (N, P, L) and necessary cofactors (M2 or p30) for RNA synthesis are colored in blue and green, respectively. The RNA-dependent RNA polymerase (RdRP) sequentially produces a gradient level of Le RNA (red line) and viral mRNAs (black, blue, or green line), with the attenuation of the downstream mRNAs at each gene junction. The Le RNA (red lines inside the box) remains uncapped and non-polyadenylated, while the viral mRNAs are 5′ capped, methylated, and 3′ polyadenylated. The lines under the Le RNA and representative viral mRNAs indicate the abundance and gradient levels of the RNA transcripts. The promoters for transcription and replication are shown with magenta arrows. (Reviewed in Liang, JVI 2020).
Figure 1: The genome organization and RNA synthesis of Mononegavirales. The negative-sense NNS genome is depicted from the 3′ end to the 5′ end, showing the 3′ leader (Le, cyan box), genes (gray, blue or green box) flanking with gene-start (GS, white box), and gene-end (GE, black box), and 5′ trailer (Tr, yellow box). The essential genes (N, P, L) and necessary cofactors (M2 or p30) for RNA synthesis are colored in blue and green, respectively. The RNA-dependent RNA polymerase (RdRP) sequentially produces a gradient level of Le RNA (red line) and viral mRNAs (black, blue, or green line), with the attenuation of the downstream mRNAs at each gene junction. The Le RNA (red lines inside the box) remains uncapped and non-polyadenylated, while the viral mRNAs are 5′ capped, methylated, and 3′ polyadenylated. The lines under the Le RNA and representative viral mRNAs indicate the abundance and gradient levels of the RNA transcripts. The promoters for transcription and replication are shown with magenta arrows. (Reviewed in Liang, JVI 2020).



 Figure 1: The genome organization and RNA synthesis of Mononegavirales. The negative-sense NNS genome is depicted from the 3′ end to the 5′ end, showing the 3′ leader (Le, cyan box), genes (gray, blue or green box) flanking with gene-start (GS, white box), and gene-end (GE, black box), and 5′ trailer (Tr, yellow box). The essential genes (N, P, L) and necessary cofactors (M2 or p30) for RNA synthesis are colored in blue and green, respectively. The RNA-dependent RNA polymerase (RdRP) sequentially produces a gradient level of Le RNA (red line) and viral mRNAs (black, blue, or green line), with the attenuation of the downstream mRNAs at each gene junction. The Le RNA (red lines inside the box) remains uncapped and non-polyadenylated, while the viral mRNAs are 5′ capped, methylated, and 3′ polyadenylated. The lines under the Le RNA and representative viral mRNAs indicate the abundance and gradient levels of the RNA transcripts. The promoters for transcription and replication are shown with magenta arrows. (Reviewed in Liang, JVI 2020).
Figure 1: The genome organization and RNA synthesis of Mononegavirales. The negative-sense NNS genome is depicted from the 3′ end to the 5′ end, showing the 3′ leader (Le, cyan box), genes (gray, blue or green box) flanking with gene-start (GS, white box), and gene-end (GE, black box), and 5′ trailer (Tr, yellow box). The essential genes (N, P, L) and necessary cofactors (M2 or p30) for RNA synthesis are colored in blue and green, respectively. The RNA-dependent RNA polymerase (RdRP) sequentially produces a gradient level of Le RNA (red line) and viral mRNAs (black, blue, or green line), with the attenuation of the downstream mRNAs at each gene junction. The Le RNA (red lines inside the box) remains uncapped and non-polyadenylated, while the viral mRNAs are 5′ capped, methylated, and 3′ polyadenylated. The lines under the Le RNA and representative viral mRNAs indicate the abundance and gradient levels of the RNA transcripts. The promoters for transcription and replication are shown with magenta arrows. (Reviewed in Liang, JVI 2020).